Wecome to ProTstab2
Stability of biomolecules, especially of proteins, is of great interest and significance. Protein stability has been the major target for protein engineering, mainly to increase the stability, but sometimes also to destabilize proteins. Effects on stability are among the most common consequences for disease-related variations, thus this phenomenon is of interest for variation interpretation to explain the effects of harmful variants. We trained a novel machine learning tool for prediction of protein stability, especially melting temperature Tm. The tool is based on amino acid sequence information and using LightGBM algorithm.
ProTstab2 was developed in collaboration between the groups of Prof. Mauno Vihinen in Lund University and Assoc. Prof. Yang Yang in Soochow University. You can also visit the mirror website in Soochow University, China.
Data sets
The dataset is 34913 proteins from 13 species. We showed 4 species'(H.sapiens, M.musculus, D.rerio, and C.elegans) distributions of melting temperature on this website. And you can see all species' distributions of melting temperature on the website: Data sets.
31470 proteins were used for method training and 3443 proteins were used for blind test. The data sets are freely available.
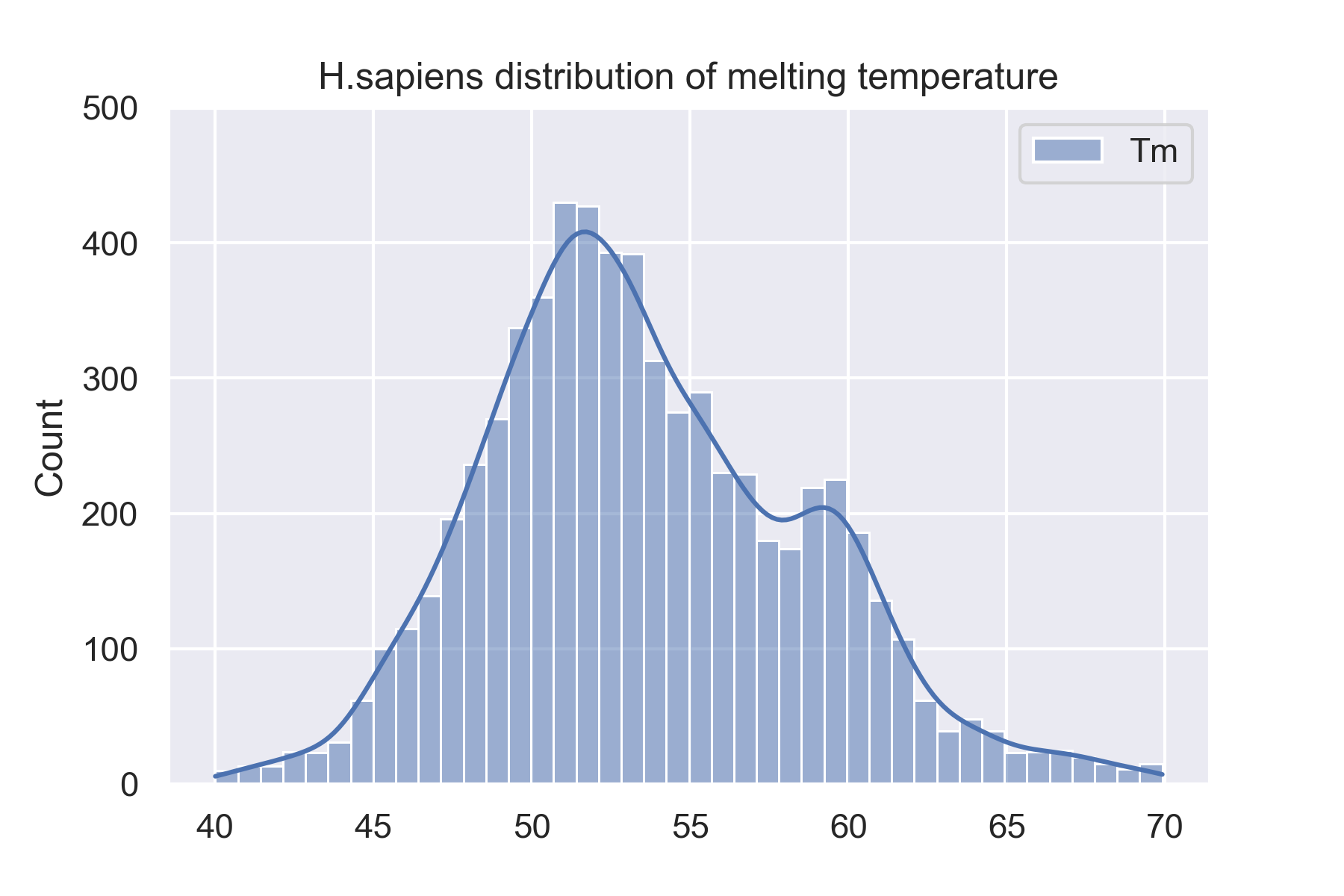
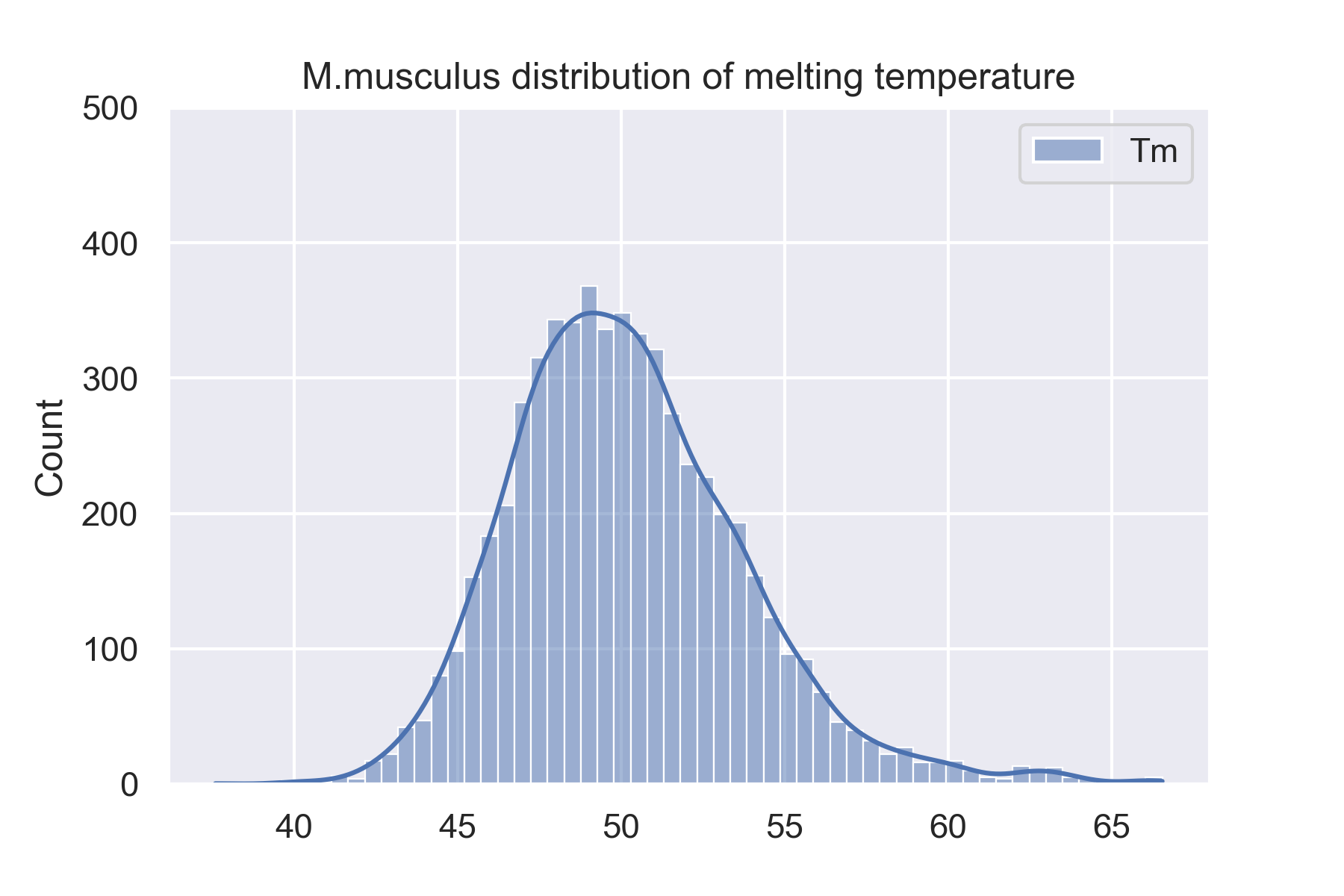
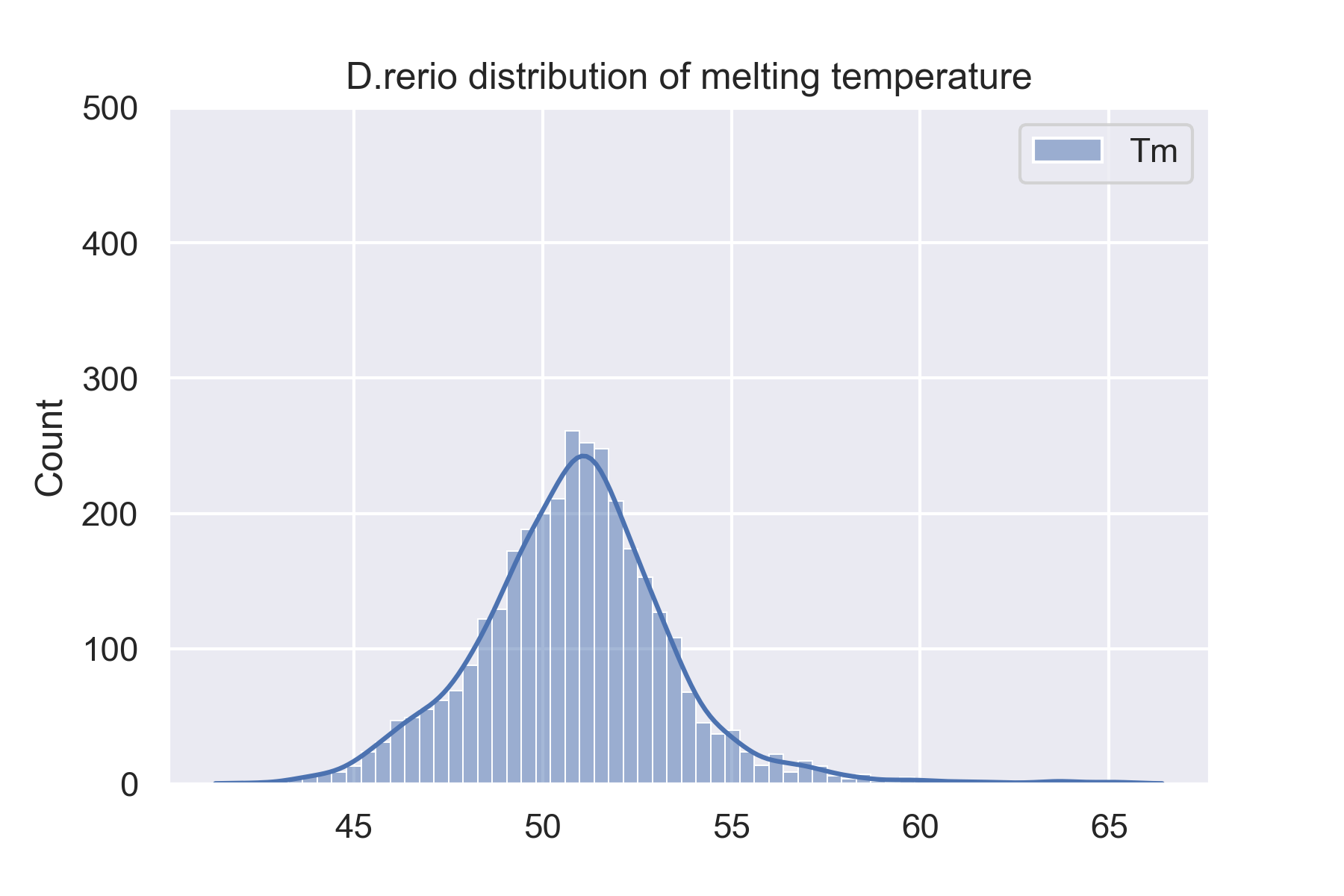
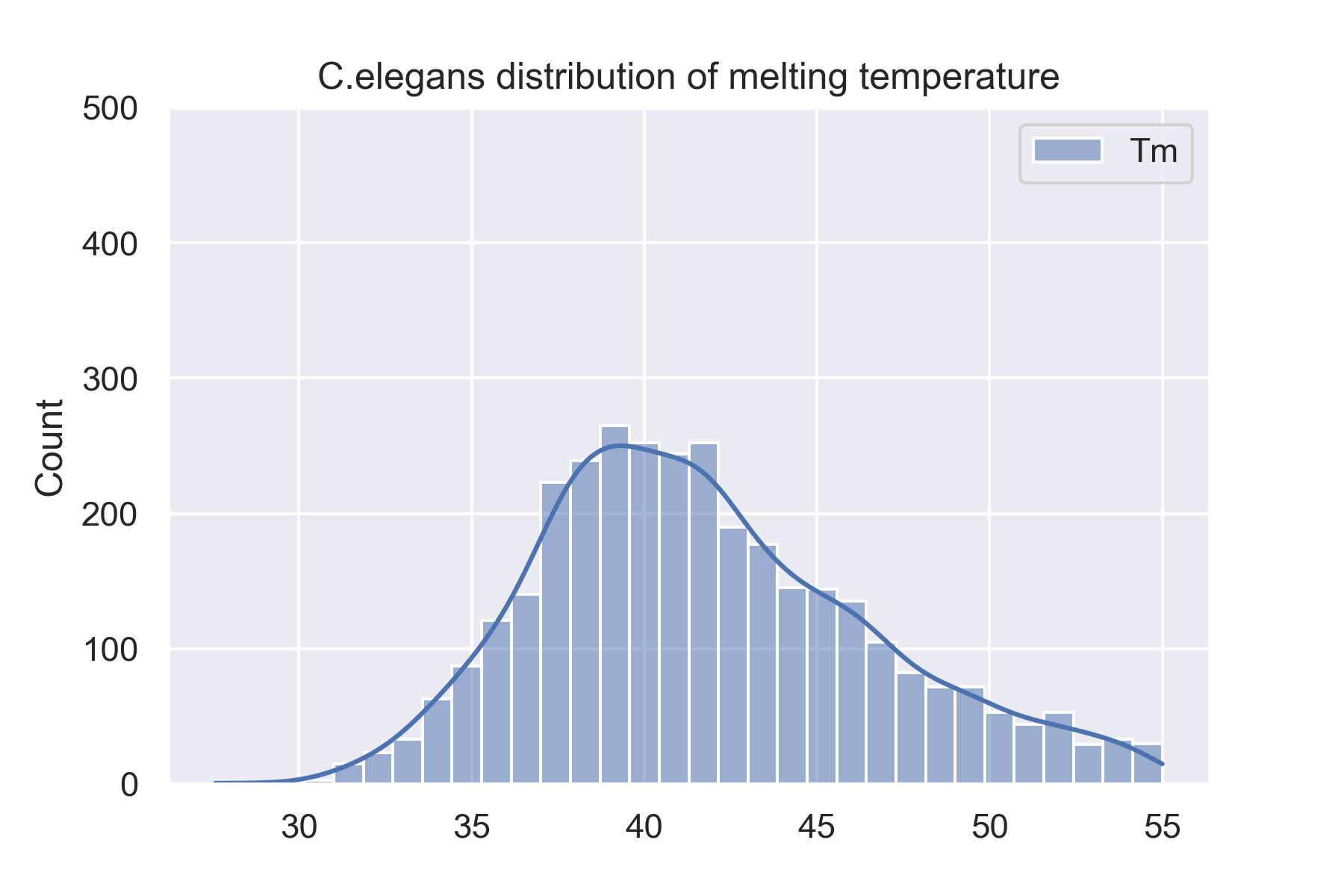
Prediction results
Result are displayed on the Results page. You can also provide your email, then the results are SENT by email.
Manuscript describing ProTstab2 is in preparation.
How to cite?
ProTstab2 was publised in
Yang Y, Zhao J, Zeng L, Vihinen M. ProTstab2 for Prediction of Protein Thermal Stabilities. Int J Mol Sci. 2022;23(18):10798. Published 2022 Sep 16.
IJMS
The original ProTstab was published in YANG Y, DING X, ZHU G, et al. ProTstab – predictor for cellular protein stability [J]. BMC Genomics, 2019, 20(1). BMC
